 Fig. 1: Functional MRI time-series.
Fig. 1: Functional MRI time-series.
 Fig. 1: Functional MRI time-series.
Fig. 1: Functional MRI time-series.
|
In recent years, a wide scope
of advanced non-invasive medical imaging techniques such as positron emission
tomography (PET), dynamic computer tomography (CT), and magnetic resonance
imaging (MRI) has been introduced into biomedical practice. Beyond the plain
imaging of morphological structure, the analysis and visualization of medical
image time-series data is a challenge with growing importance for both basic
research and clinical application.
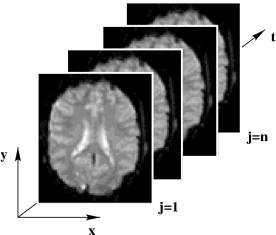
Fig. 1 shows an example of an image time-series generated from functional magnetic resonance imaging (fMRI). For each voxel, we obtain a signal time-series that may be interpreted in the light of the experimental conditions. The visual interpretation of such spatiotemporal data sets on the part of human beings is limited by physical dimensions with only two-dimensional, or pseudo-three-dimensional data plotting methods. In our group, we explore innovative algorithms that allow multidimensional dynamic biomedical data sets to be compressed into low-dimensional representations for visual analysis, such as projection and clustering methods. |
 |
| Fig. 2: Cluster assignment maps for fMRI. |
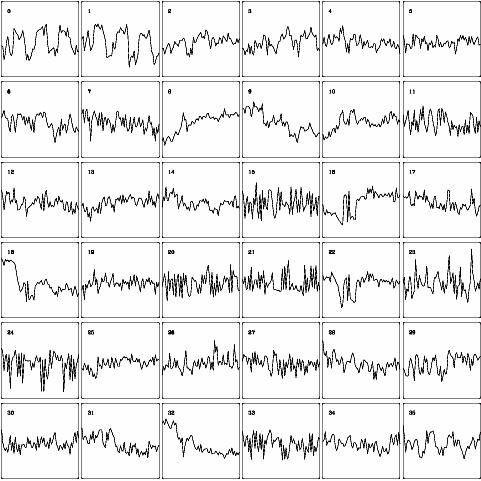 |
| Fig. 3: Corresponding time-series for fig. 2 |
|
Figs. 2 and 3 show examples for the
analysis of fMRI time-series obtained by a method called minimal free energy
vector quantization. This clustering procedure identifies groups of pixels
sharing similar properties of signal dynamics. These can be interpreted in the
light of physiological knowledge about the experiment. Clusters #0 and #1 in
fig. 2, for example, represent activated regions of the visual cortex in a
photic stimulation experiment. The corresponding time-series reflect the shape
of the stimulus paradigm.
This data visualization strategy can be helpful for other biomedical image time-series as well, such as magnetic resonance mammography data for the analysis of suspicious lesions in patients with breast cancer, or time-series from patient examinations in nuclear medicine. Although there are obvious methodological similarities, each clinical application requires specific careful consideration with regard to data preprocessing, postprocessing, and interpretation. This challenge can only be managed by close interdisciplinary co-operation of medical doctors, engineers, and computer scientists. Hence, this field of research can serve as an example of lively cross-fertilization between theoretical research on exploratory data analysis and practical application in clinical medicine. |
|
Last Update: March-20-2002 |